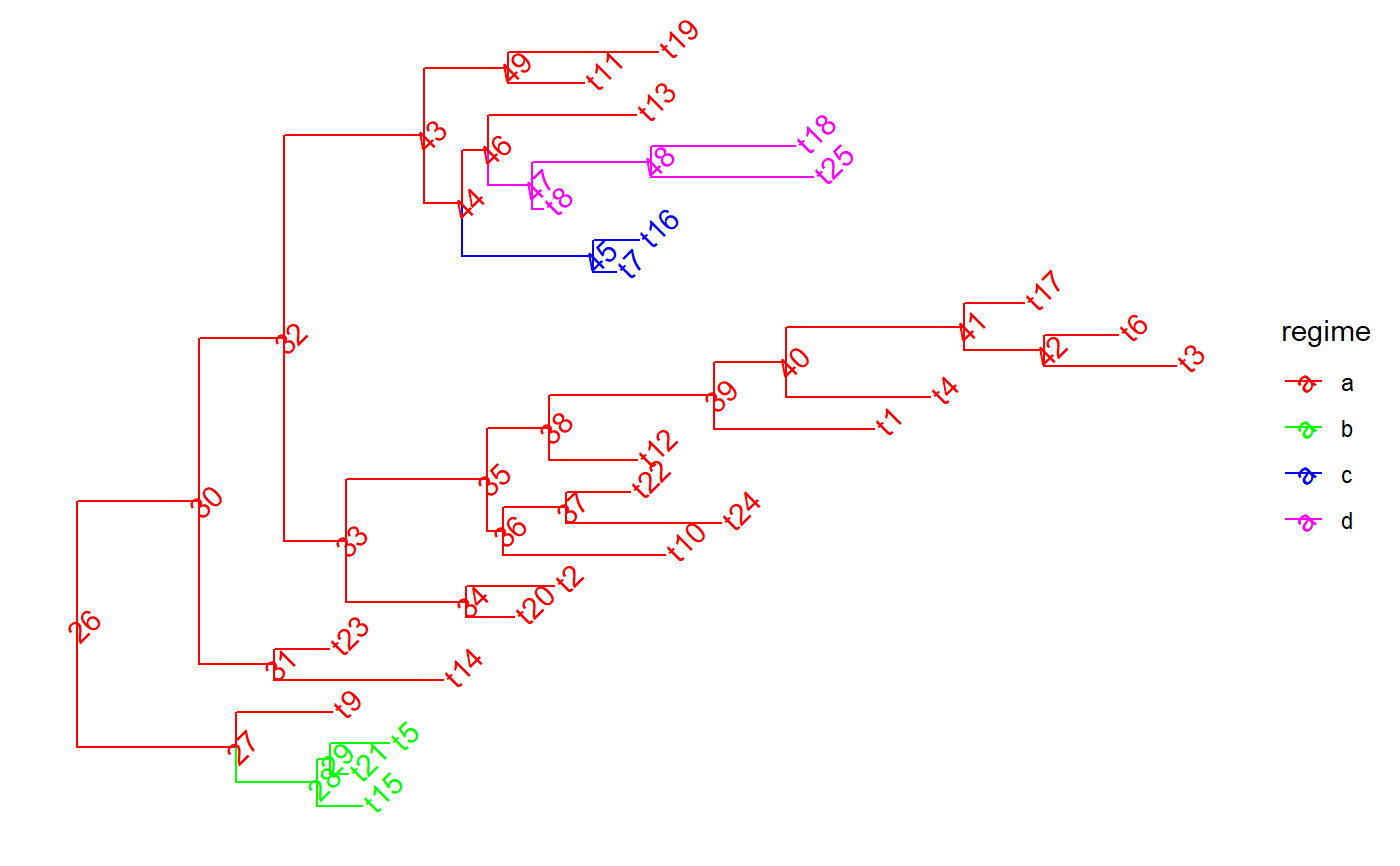
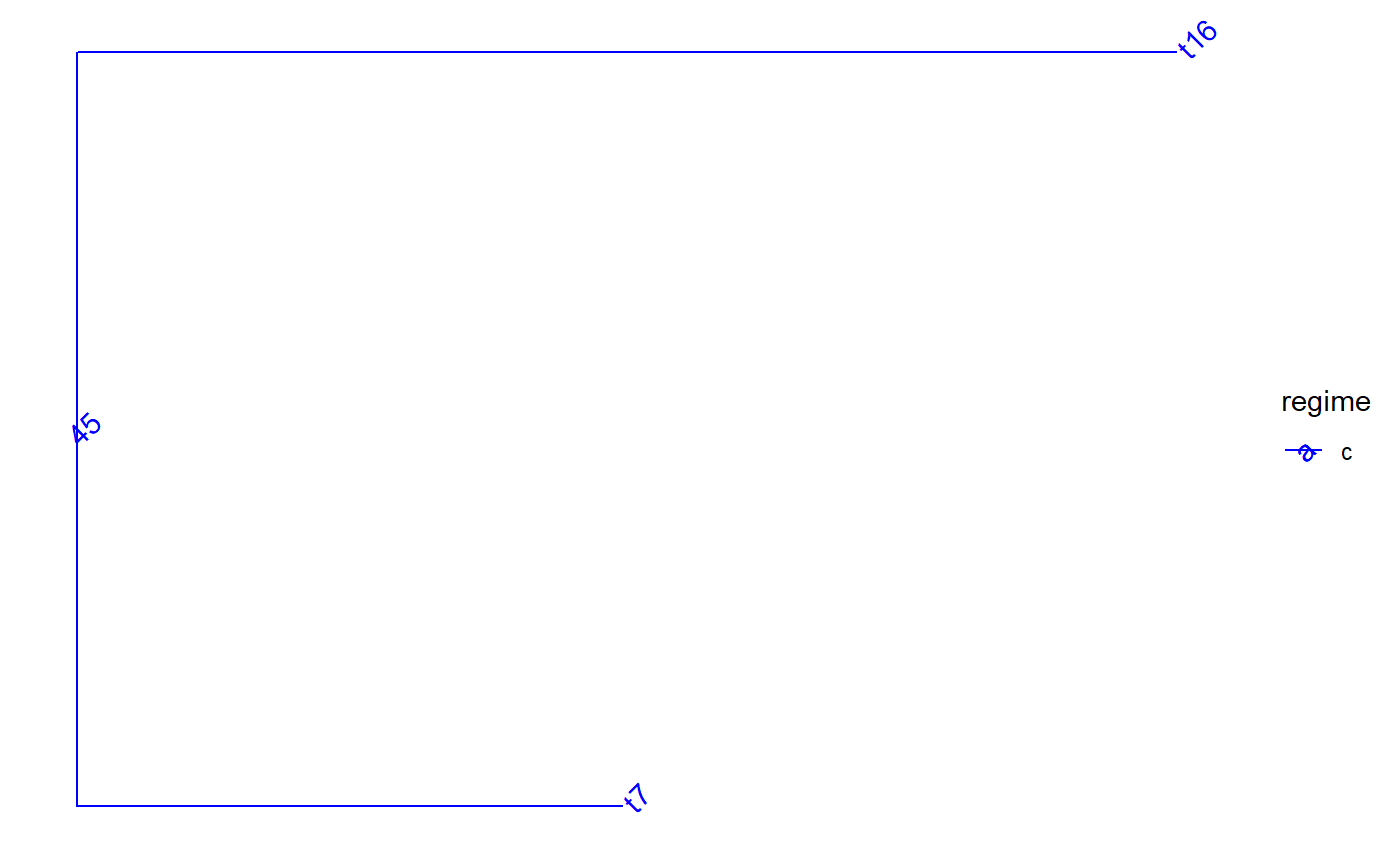
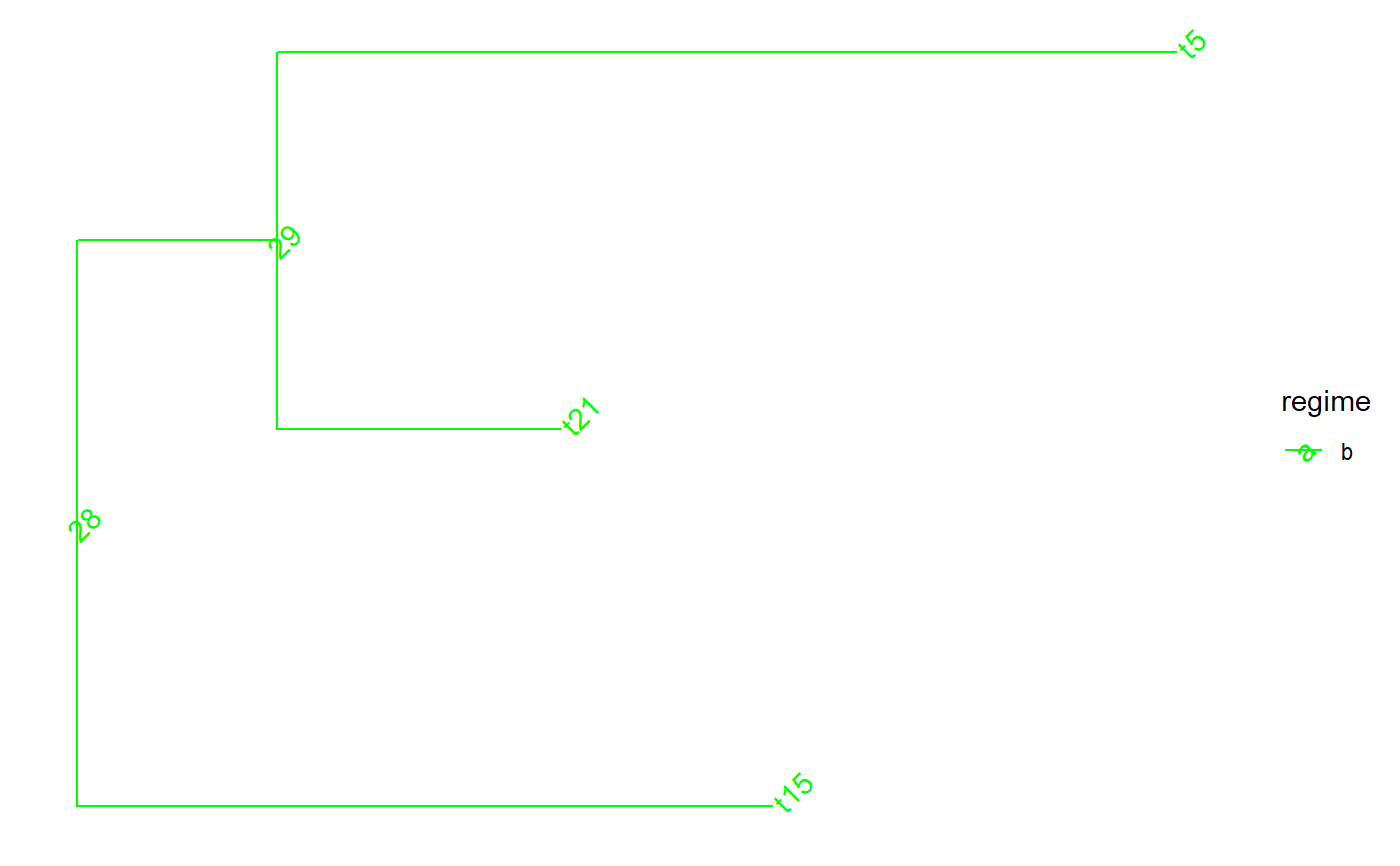
Perfrorm nested extractions or drops of clades from a tree
PCMTreeEvalNestedEDxOnTree.RdPerfrorm nested extractions or drops of clades from a tree
PCMTreeEvalNestedEDxOnTree(expr, tree)
Arguments
| expr | a character string representing an R expression of nested calls
of functions
|
|---|---|
| tree | a phylo object with named tips and internal nodes |
Value
the resulting phylo object from evaluating expr on tree.
Examples
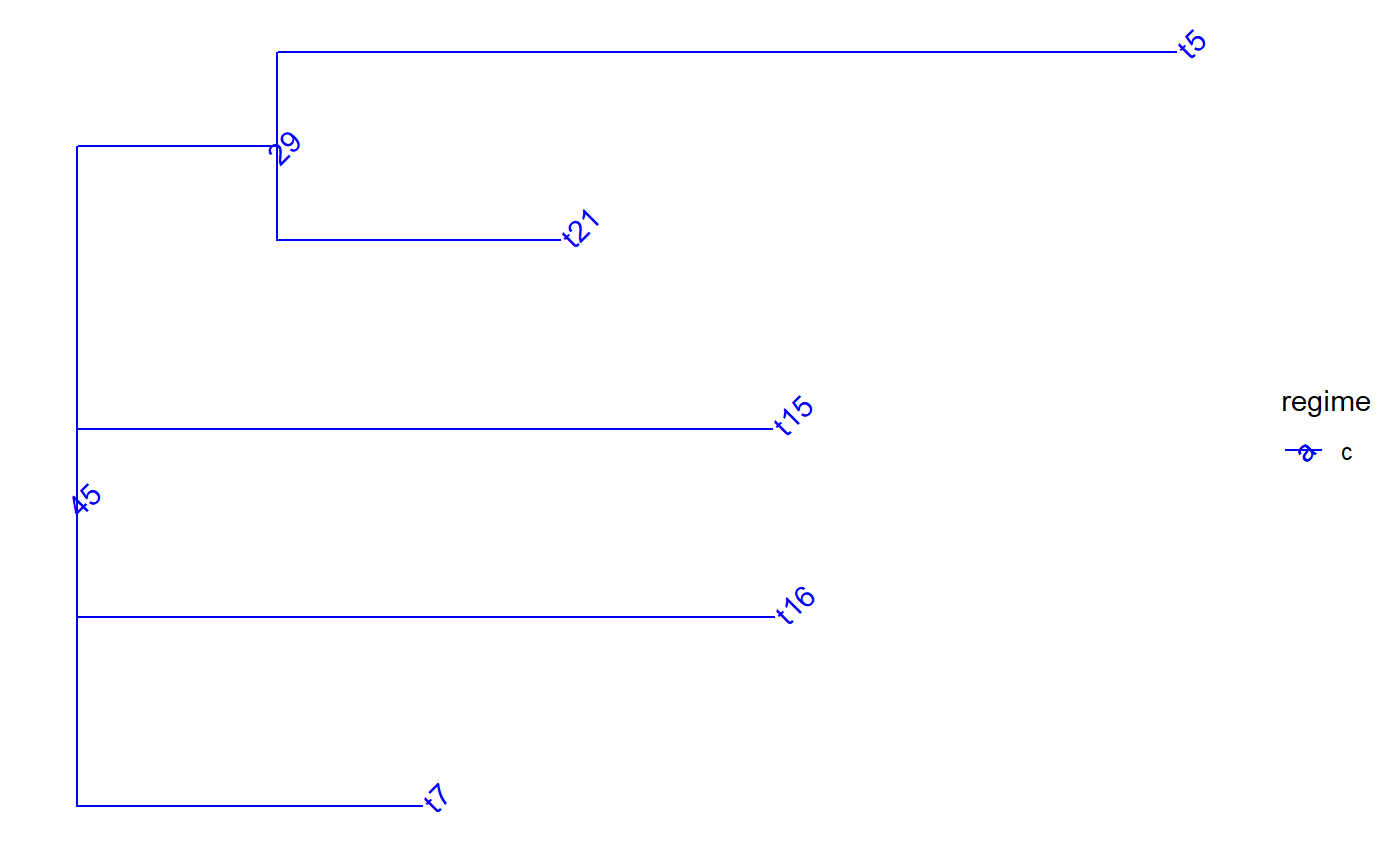
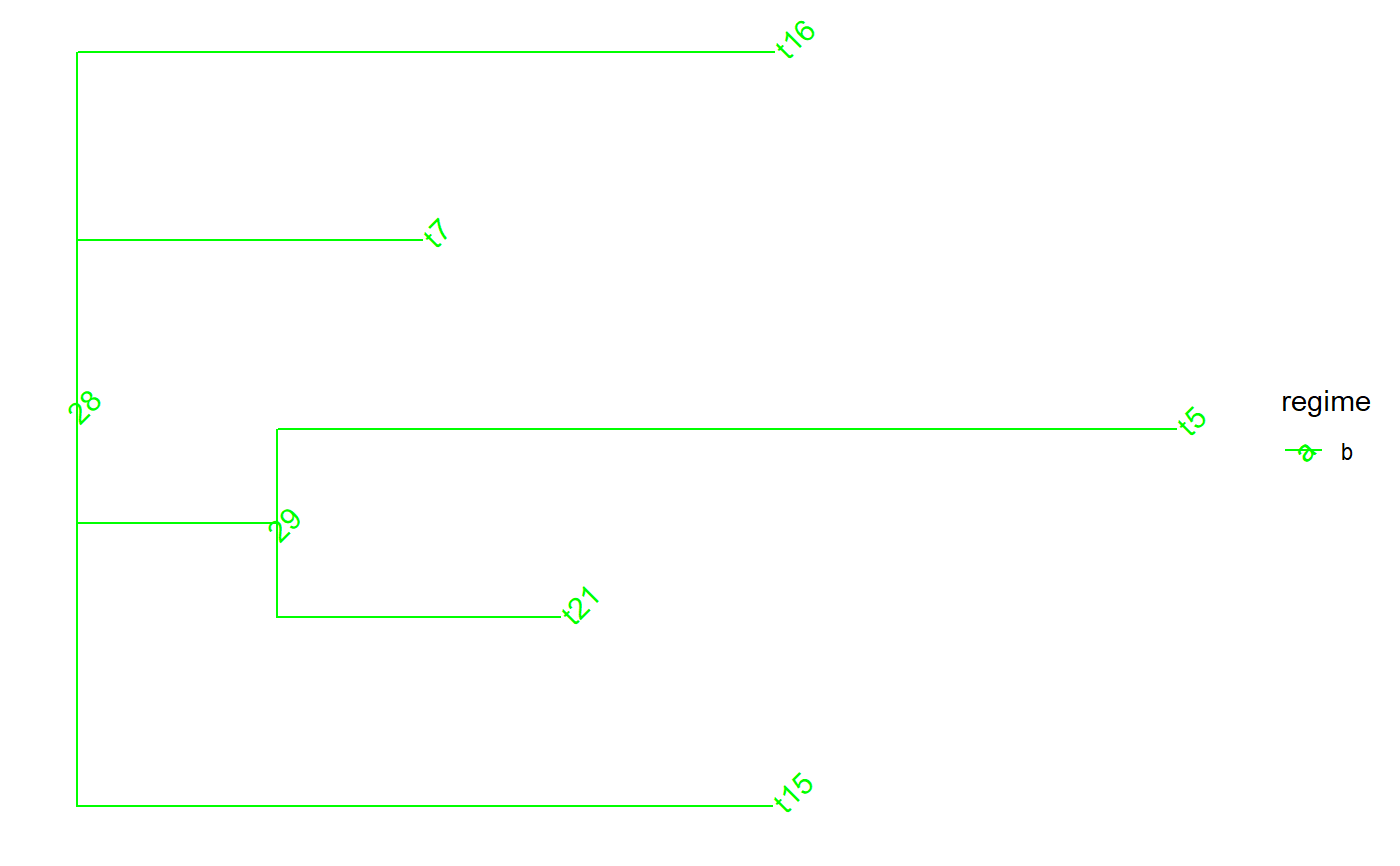
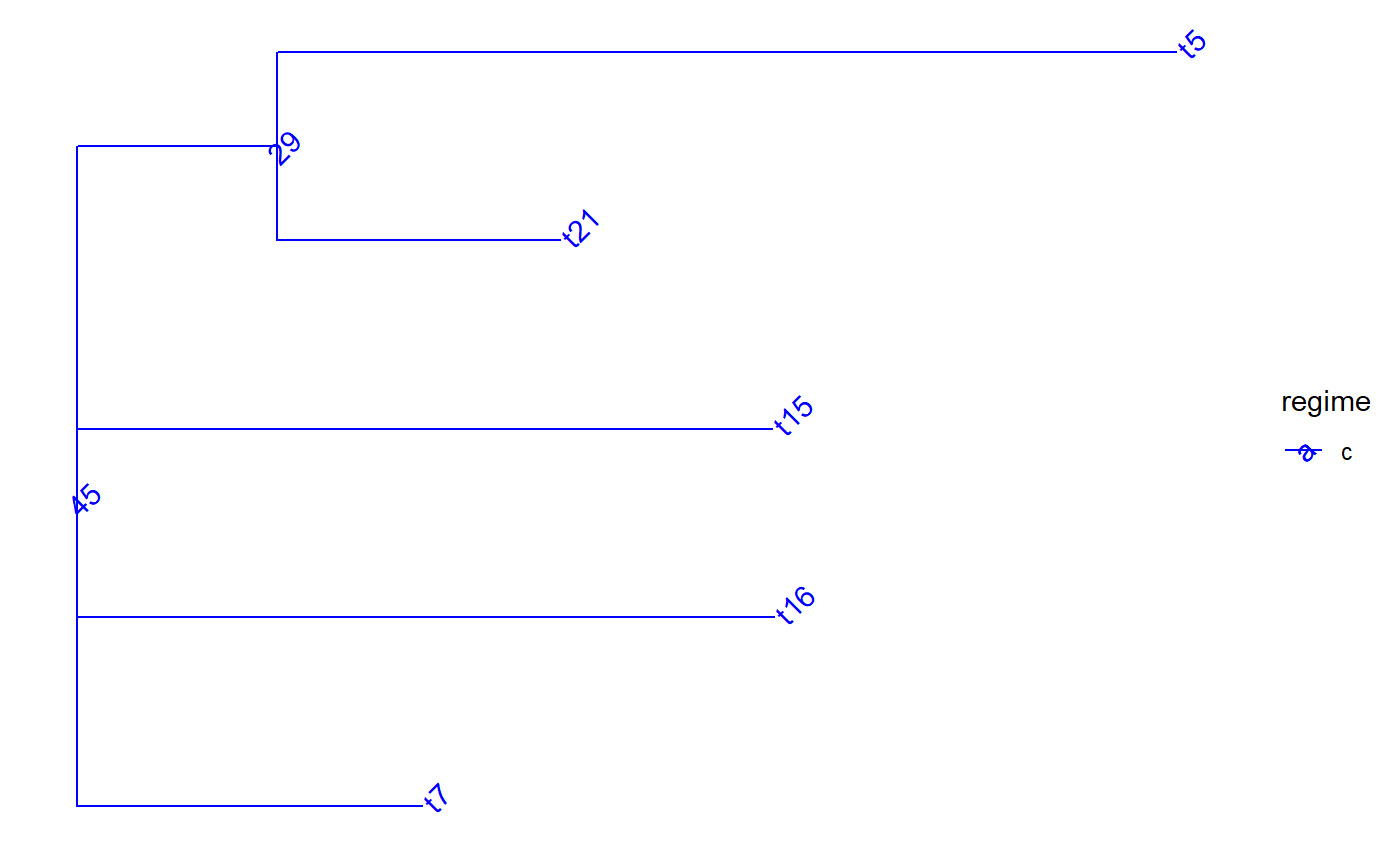
set.seed(1, kind = "Mersenne-Twister", normal.kind = "Inversion") tree <- PCMTree(ape::rtree(25)) PCMTreeSetPartRegimes( tree, c(`26`="a", `28`="b", `45`="c", `47`="d"), setPartition = TRUE) # \donttest{ PCMTreePlot( tree, palette=c(a = "red", b = "green", c = "blue", d = "magenta")) + ggtree::geom_nodelab(angle = 45) + ggtree::geom_tiplab(angle = 45)# } bluePart <- PCMTreeEvalNestedEDxOnTree("D(E(tree,45),47)", tree) PCMTreeGetPartRegimes(bluePart)#> 45 #> "c"# \donttest{ PCMTreePlot( bluePart, palette=c(a = "red", b = "green", c = "blue", d = "magenta")) + ggtree::geom_nodelab(angle = 45) + ggtree::geom_tiplab(angle = 45)# } # Swapping the D and E calls has the same result: bluePart2 <- PCMTreeEvalNestedEDxOnTree("E(D(tree,47),45)", tree) PCMTreeGetPartRegimes(bluePart2)#> 45 #> "c"# \donttest{ PCMTreePlot( bluePart2, palette=c(a = "red", b = "green", c = "blue", d = "magenta")) + ggtree::geom_nodelab(angle = 45) + ggtree::geom_tiplab(angle = 45)# } greenPart <- PCMTreeEvalNestedEDxOnTree("E(tree,28)", tree) bgParts <- bluePart+greenPart # \donttest{ PCMTreePlot( greenPart, palette=c(a = "red", b = "green", c = "blue", d = "magenta")) + ggtree::geom_nodelab(angle = 45) + ggtree::geom_tiplab(angle = 45)PCMTreePlot( bluePart + greenPart, palette=c(a = "red", b = "green", c = "blue", d = "magenta")) + ggtree::geom_nodelab(angle = 45) + ggtree::geom_tiplab(angle = 45)PCMTreePlot( greenPart + bluePart, palette=c(a = "red", b = "green", c = "blue", d = "magenta")) + ggtree::geom_nodelab(angle = 45) + ggtree::geom_tiplab(angle = 45)PCMTreePlot( bgParts, palette=c(a = "red", b = "green", c = "blue", d = "magenta")) + ggtree::geom_nodelab(angle = 45) + ggtree::geom_tiplab(angle = 45)# }