Extract a clade from phylogenetic tree
PCMTreeExtractClade.RdExtract a clade from phylogenetic tree
PCMTreeExtractClade( tree, cladeRootNode, tableAncestors = NULL, X = NULL, returnList = !is.null(X) )
Arguments
| tree | a PCMTree object. |
|---|---|
| cladeRootNode | a character string denoting the label or an integer denoting a node in the tree. |
| tableAncestors | an integer matrix returned by a previous call to PCMTreeTableAncestors(tree) or NULL. |
| X | an optional k x N matrix with trait value vectors for each tip in tree. |
| returnList | logical indicating if only the phylo object associated
with the clade should be returned. Defaults to |
Value
If returnList is FALSE, a phylo object associated with the clade, otherwise, a list with two named members :
treethe phylo object associated with the clade
Xthe submatrix of X with columns corresponding to the tips in the clade
See also
PCMTreeSpliAtNode PCMTreeDropClade
Examples
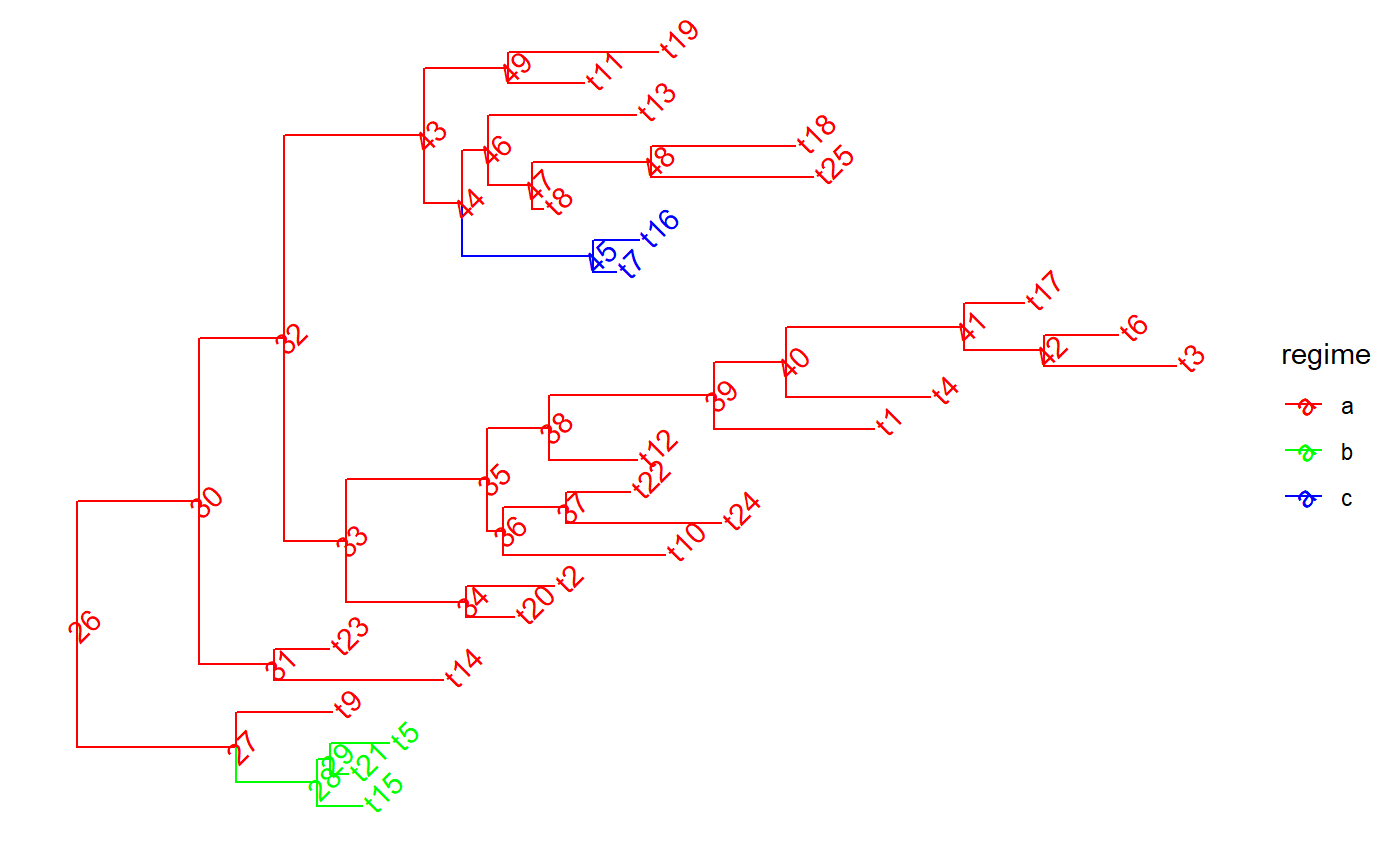
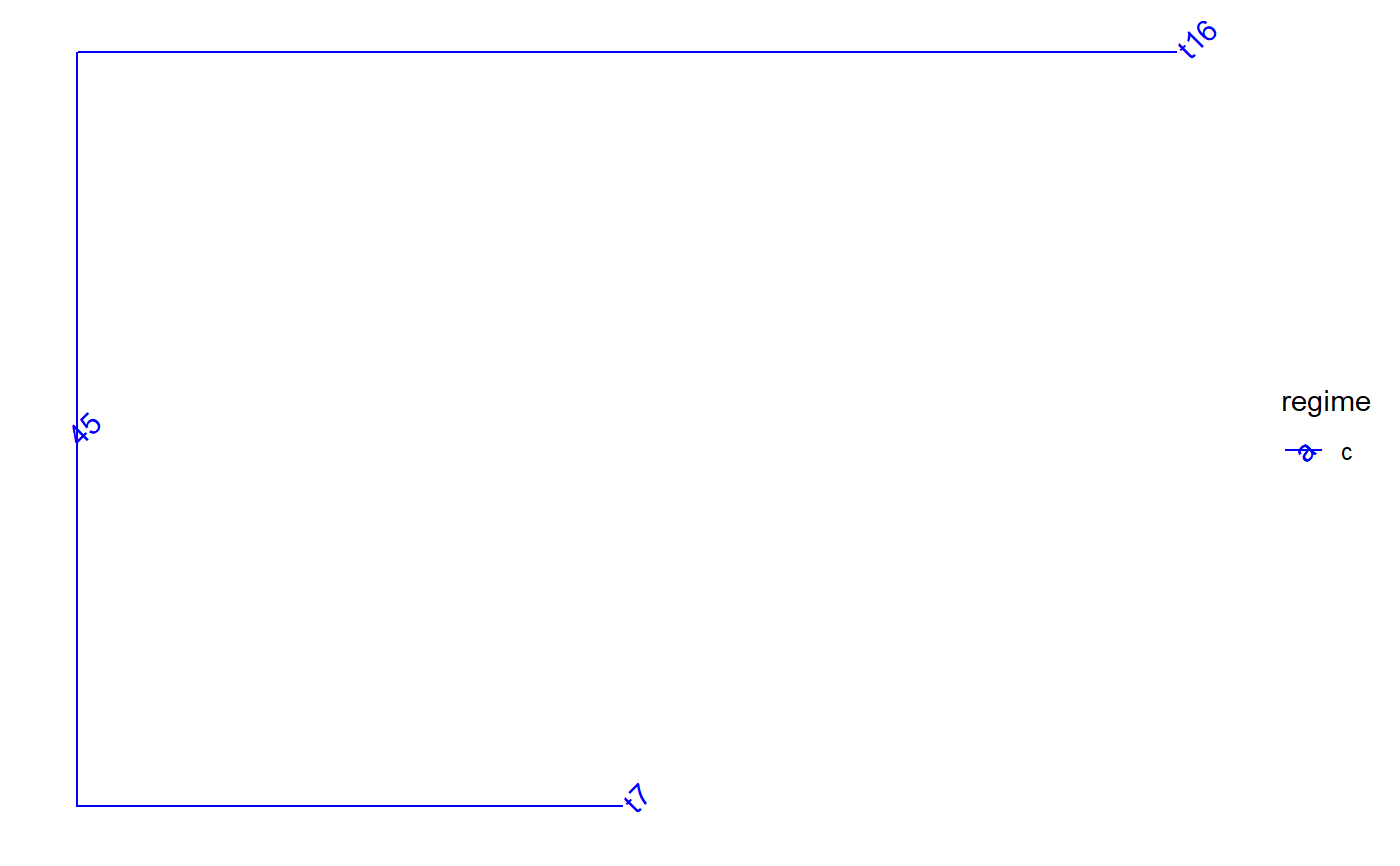
set.seed(1, kind = "Mersenne-Twister", normal.kind = "Inversion") tree <- PCMTree(ape::rtree(25)) PCMTreeSetPartRegimes( tree, c(`26`="a", `28`="b", `45`="c"), setPartition = TRUE) # \donttest{ PCMTreePlot(tree, palette=c(a = "red", b = "green", c = "blue")) + ggtree::geom_nodelab(angle = 45) + ggtree::geom_tiplab(angle = 45)#> 45 #> "c"# \donttest{ PCMTreePlot(blueTree, palette=c(a = "red", b = "green", c = "blue")) + ggtree::geom_nodelab(angle = 45) + ggtree::geom_tiplab(angle = 45)# } # we need to use the label here, because the node 29 in tree is not the same # id in redGreenTree: blueTree2 <- PCMTreeDropClade(blueTree, "48") # \donttest{ PCMTreePlot(blueTree2, palette=c(a = "red", b = "green", c = "blue")) + ggtree::geom_nodelab(angle = 45) + ggtree::geom_tiplab(angle = 45)# }